Abstract
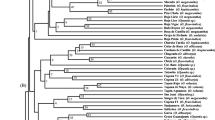
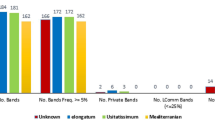
A genetic variability analysis involving 45 accessions of Macadamia including four species, M. integrifolia, M. tetraphylla, M. ternifolia, and M. hildebrandii and a wild relative, Hicksbeachia pinnatifolia was performed using eight enzyme systems encoded by 16 loci (Gpi-1 and 2, Idh-1 and 2, Lap, Mdh-1, 2, and 3, 6Pgd-2, Pgm-2 and 3, Tpi-1 and 2, Ugpp-1, 2, and 3). Forty-three accessions possessed distinct isozyme fingerprints indicating a high level of genetic variation. Examination of multivariate relationships among accessions using a cluster analysis resulted in the identification of four clusters, two of which contained one accession each representing H. pinnatifolia and M. hildebrandii. All Hawaiian cultivars were included in two sub-clusters within the largest cluster, which encompassed all the M. integrifolia and three M. ternifolia accessions, suggesting that: (1) the Hawaiian cultivars must have originated from at least two genetically diverse ancestral populations and (2) M. ternifolia may be a conspecific variant of M. integrifolia. Macadamia tetraphylla has diverged marginally from the M. integrifolia and M. ternifolia complex suggesting that these taxa represent a species complex. The different measures of genetic variability such as mean number of alleles per locus, polymorphic index, and observed and expected levels of heterozygosity, indicated significant levels of genetic variation in the Macadamia collection.
Similar content being viewed by others
References
Aradhya, K.M., F.T. Zee & R.M. Manshardt, 1994. Isozyme variation in cultivated and wild pineapple. Euphytica 79: 87–99.
Aradhya, K.M., F.T. Zee & R.M. Manshardt, 1995. Isozyme variation in lychee (Litchi chinensis Sonn.). Scientia Horticulturae 63: 21–35.
Arulsekar, S. & D.E. Parfitt, 1986. Isozyme analysis procedures for stone fruits, almond, grape, walnut, pistachio, and fig. HortScience 21: 928–933.
Beaumont, J.H., 1958. The Macadamia in Australia and Hawaii. California Mac. Soc. Year Book 4: 25–29.
Francis, W.D., 1951. Australian rain forest trees. Angus & Robertson, Sydney, 455 pp.
Hamilton, R.A. & E.T. Fukunaga, 1959. Mcadamia nuts in Hawaii. Hawaii Agril. Experimental Station Bulletin 121, University of Hawaii, Honolulu, Hawaii, 51 pp.
Hamilton, R.A. & P.J. Ito, 1984. Macadamia nut cultivars recommended for Hawaii. Info. Text Series 023, Hawaii Institute of Tropical Agriculture and Human Resources, University of Hawaii, Honolulu, Hawaii, 7 pp.
Hamrick, J.L. & M.J. Godt, 1990. Allozyme diversity in plant species. In Brown, H.D., M.T. Clegg, A.L. Kahler & B.S. Weir (Eds.), Plant population genetics, breeding, and genetic resources. Sinauer Associates Inc., Sunderland, Massachusetts, pp. 43–63.
Hawaii Agricultural Statistical Service, 1996. Macadamia nuts, production and value, Final estimates, 1995–96. Hawaii Agricultural Statistical Service, P.O. Box 22159, Honolulu, Hawaii, 4 pp.
Nei, M., 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–90.
Rohlf, F.J. & R.R. Sokal, 1981. Comparing numerical taxonomic studies. Syst. Zool. 30: 459–90.
Shaw, C.R.&R. Prasad, 1970. Starch gel electrophoresis of enzymes – a compilation of recipies. Biochemical Genetics 4: 297–320.
Shigeura, G.T.& H. Ooka, 1984. Macadamia Nuts in Hawaii: History and Production. Res. Extn. Series 039, Hawaii Institute ofTropical Agriculture and Human Resources, University of Hawaii, Honolulu, 91 pp.
Smith, L.S., 1956. New species and notes on Queensland plants. Proc. Royal Soc. Queensland 67: 29–35.
Sokal, R.R. & C.D. Michener, 1958. A statistical method for evaluating systematic relationships. Univ. ofKansas Scientific Bulletin 38: 1409–38.
Soltis, D.E., C.H. Haufler, D.C. Darrow & G.J. Gastony, 1983. Starch gel electrophoresis of ferns: A compilation of grinding buffers, gel and electrode buffers, and staining schedules. American Fern Journal 73: 9–27.
Stephenson, R.A. & C.W. Winks, 1992. Macadamia integrifolia Maiden & Betche. In Verheij, E.W.M. & R.E. Coronel (Eds.), Plant Resources of South-East Asia 2. Edible fruits and nuts. PROSEA, Bogor Indonesia, pp. 195–198.
Sui, L., F. Zee, R.M. Manshardt & M.K. Aradhya, 1995. Enzyme polymorphisms inCanarium. ScientiaHorticulturae 68: 197–206.
Van Steenis, C.G.G.J., 1952. Miscellaneous botanical notes. Ic. Reinwardtia 1: 467–81.
Vithanage, V. & C.W. Winks, 1992. Isozymes as genetic markers for Macadamia. Scientia Horticulturae 49: 103–115.
Weeden, N.F. & J.F. Wendel, 1989. Genetics of plant isozymes. In Soltis, D.E. & P.S. Soltis (Eds.), Isozymes in plant biology. Dioscorides Press, Portland, pp. 46–72.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Aradhya, M.K., Yee, L.K., Zee, F.T. et al. Genetic variability in Macadamia. Genetic Resources and Crop Evolution 45, 19–32 (1998). https://doi.org/10.1023/A:1008634103954
Issue Date:
DOI: https://doi.org/10.1023/A:1008634103954