Abstract
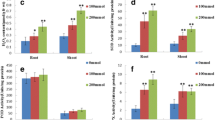
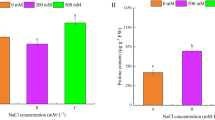
Knowledge of stress-responsive proteins is critical for further understanding the molecular mechanisms of stress tolerance. The objectives of this study were to establish a proteomic map for a perennial grass species, creeping bentgrass (A. stolonifera L.), and to identify differentially expressed, salt-responsive proteins in two cultivars differing in salinity tolerance. Plants of two cultivars (‘Penncross’ and ‘Penn-A4’) were irrigated daily with water (control) or NaCl solution to induce salinity stress in a growth chamber. Salinity stress was obtained by adding NaCl solution of 2, 4, 6, and 8 dS m−1 in the soil daily for 2-day intervals at each concentration, and then by watering soil with 10 dS m−1 solution daily for 28 days. For proteomic map, using two-dimensional electrophoresis (2-DE), approximately 420 and 300 protein spots were detected in leaves and roots, respectively. A total of 148 leaf protein spots and 40 root protein spots were excised from the 2-DE gels and subjected to mass spectrometry analysis. In total, 106 leaf protein spots and 24 root protein spots were successfully identified. Leaves had more salt-responsive proteins than roots in both cultivars. The superior salt tolerance in ‘Penn-A4’, indicated by shoot extension rate, relative water content, and cell membrane stability during the 28-day salinity stress could be mainly associated with its higher level of vacuolar H+-ATPase in roots and UDP-sulfoquinovose synthase, methionine synthase, and glucan exohydrolase in leaves, as well as increased accumulation of catalase and glutathione S-transferase in leaves. Our results suggest that salinity tolerance in creeping bentgrass could be in part controlled by an alteration of ion transport through vacuolar H+-ATPase in roots, maintenance of the functionality and integrity of thylakoid membranes, sustained polyamine biosynthesis, and by the activation of cell wall loosening proteins and antioxidant defense mechanisms.




Similar content being viewed by others
References
Alves M, Francisco R, Martins I, Ricardo CPP (2006) Analysis of Lupinus albus leaf apoplastic proteins in response to boron deficiency. Plant Soil 279:1–11
Askari H, Edqvist J, Hajheidari M, Kafi M, Salekdeh GH (2006) Effects of salinity levels on proteome of Suaeda aegyptiaca leaves. Proteomics 5:2542–2554
Baek D, Jin Y, Jeong JC, Lee H, Moon H et al (2008) Suppression of reactive oxygen species by glyceraldehyde-3-phosphate dehydrogenase. Phytochemistry 69:333–338
Bernstein N, Meiri A, Zilberstaine M (2004) Root growth of avocado [Persea americana Mill] is more sensitive to salinity than shoot growth. J Am Soc Hortic Sci 129:188–192
Bevan M, Bancroft I, Bent E, Love K et al (1998) Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391:485–488
Bradford MM (1976) A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye-binding. Anal Biochem 72:248–254
Carrow RN, Duncan RR (1998) Salt-affected turfgrass sites: assessment and management. Wiley, New York
Chang W, Huang L, Shen M, Webster C et al (2000) Patterns of protein synthesis and tolerance of anoxia in root tips of maize seedlings acclimated to a low-oxygen environment and identification of proteins by mass spectrometry. Plant Physiol 122:295–317
Chinnusamy V, Jagendorf A, Zhu JK (2005) Understanding and improving salt tolerance in plants. Crop Sci 45:437–448
Chitteti BR, Peng Z (2007) Proteome and phosphoproteome differential expression under salinity stress in rice (Oryza sativa) roots. J Proteome Res 6:1718–1727
Dixon DP, Lapthorn A, Edwards R (2002) Plant glutathione transferases. Genome Biol 3:3004.1–3004.10
Dooki AD, Mayer-Posner FJ, Askari H, Zaiee A, Salekdeh GH (2006) Proteomic responses of rice young panicles to salinity. Proteomics 6:6498–6507
Duclos-Vallee JC, Capel F, Mabit H, Petit MA (1998) Phosphorylation of the hepatitis B virus core protein by glyceraldehyde-3-phosphate dehydrogenase protein kinase activity. J Gen Virol 79:1665–1670
Edwards R, Dixon D, Walbot V (2000) Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci 5:193–198
Engel M, Seifert M, Theisinger B, Seyfert U, Welter C (1998) Glyceraldehyde-3-phosphate dehydrogenase and Nm23-H1/nucleoside diphosphate kinase A: two old enzymes combine for the novel Nm23 protein phosphotransferase function. J Biol Chem 273:20058–20065
Evans PT, Malmberg RL (1989) Do polyamines have roles in plant development? Annu Rev Plant Physiol Plant Mol Biol 40:235–269
Ferreira S, Hjernø K, Larsen M, Wingsle G et al (2006) Proteome profiling of Populus euphratica Oliv. upon heat stress. Ann Bot 98:361–377
Flores HE, Protacio CM, Signs MW (1989) Primary and secondary metabolism of polyamines in plants. In: Poulton JE, Romeo JT, Conn EE (eds) Plant nitrogen metabolism. Recent Advances Phytochemistry, vol 23. Plenum Press, New York, pp 329–393
Galston AW, Sawhney RK (1990) Polyamines in plant physiology. Plant Physiol 94:406–410
Gazanchian A, Hajheidari M, Sima NK, Salekdeh GH (2007) Proteome response of Elymus elongatum to severe water stress and recovery. J Exp Bot 58:291–300
Georgopoulos C, Welch WJ (1993) Role of the major heat shock proteins as molecular chaperones. Annu Rev Cell Biol 9:601–634
Giavalisco P, Nordhoff E, Kreitler T, Kloppel K et al (2005) Proteome analysis of Arabidopsis thaliana by two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionisation-time of flight mass spectrometry. Proteomics 5:1902–1913
Green L, Yee B, Buchanan B, Kamide K et al (1991) Ferredoxin and ferredoxin-nadp reductase from photosynthetic and nonphotosynthetic tissues of tomato. Plant Physiol 96:1207–1213
Hajheidari M, Salekdeh GH, Heidari M, Abdollahian-Noghabi M, Sadeghian SY (2005) Proteome analysis of sugar beet leaves under drought stress. Proteomics 5:950–960
Hajheidari M, Eivazi A, Buchanan BB, Wong JH et al (2007) Proteomics uncovers a role for redox in drought tolerance in wheat. J Proteome Res 6:1451–1460
Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51:463–499
Hiraga S, Sasaki K, Ito H, Ohashi Y, Matsui H (2001) A large family of class III plant peroxidases. Plant Cell Physiol 42:462–468
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. California Agric Exp Stn Circ 347:1–32
Hrmova M, Harvey AJ, Wang J, Shirley NJ et al (1996) Barley β-d-glucan exohydrolases with β-d-glucosidase activity. J Biol Chem 271:5277–5286
Imin N, Kerim T, Weinman JJ, Rolfe BG (2004) Effect of early cold stress on the maturation of rice anthers. Proteomics 4:1873–1882
Jiang Y, Huang B (2002) Protein alterations in tall fescue in response to drought stress and abscisic acid. Crop Sci 42:202–207
Jiang Y, Yang B, Harris NS, Deyholos MK (2007) Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J Exp Bot 58:3591–3607
Kopriva S, Bauwe H (1995) Serine hydroxymethyltransferase from Solanum tuberosum. Plant Physiol 107:271–272
Koussevitzky S, Suzuki N, Huntington S, Armijo L et al (2008) Ascorbate peroxidase 1 plays a key role in the response of Arabidopsis thaliana to stress combination. J Biol Chem 283:34197–34203
Labra M, Gianazza E, Waitt R, Eberini I et al (2006) Zea mays L. protein changes in response to potassium dichromate treatments. Chemosphere 62:1234–1244
Lebherz HG, Leadbetter MM, Bradshaw RA (1984) Isolation and characterization of the cytosolic and chloroplast form of spinach leaf fructose diphosphate aldolase. J Biol Chem 259:1011–1017
Lee G, Carrow RN, Duncan RR (2004) Photosynthetic responses to salinity stress of halophytic seashore paspalum ecotypes. Plant Sci 166:1417–1425
Lee DG, Ahsan N, Lee S, Kang KY et al (2007) A proteomic approach in analyzing heat-responsive proteins in rice leaves. Proteomics 7:3369–3383
Marcum KB, Murdoch CL (1990) Growth responses, ion relations, and osmotic adaptations of eleven C4 turfgrasses to salinity. Agron J 82:892–896
Messiaen J, Cambier P, Cutsem P (1997) Polyamines and pectins I. Ion exchange and selectivity. Plant Physiol 113:387–395
Moon H, Lee B, Choi G, Shin D et al (2003) NDP kinase interacts with two oxidative stress-activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proc Natl Acad Sci USA 100:358–363
Mooney BP, Miernyka JA, Greenlief CM, Thelena JJ (2006) Using quantitative proteomics of Arabidopsis roots and leaves to predict metabolic activity. Physiol Plant 128:237–250
Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25:239–250
Nagy E, Rigby WFC (1995) Glyceraldehyde-3-phosphate dehydrogenase selectively binds Au-rich RNA in the NAD+ -binding region (Rossmann fold). J Biol Chem 270:2755–2769
Ndimba BK, Chivasa S, Simon WJ, Slabas AR (2005) Identification of Arabidopsis salt and osmotic stress responsive proteins using two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 5:4185–4196
Neuhoff V, Arold N, Taube D, Ehrhardt D (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9:255–262
Noctor G, Foyer CH (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49:249–279
Orcutt DM, Nilsen ET (2000) The physiology of plants under stress: soil and biotic factors. Wiley, New York, pp 177–236
Passardi F, Cosio C, Penel C, Dunand C (2005) Peroxidases have more functions than a Swiss army knife. Plant Cell Rep 24:255–265
Pessarakli M, Touchane H (2006) Growth responses of bermudagrass and seashore paspalum under various levels of sodium chloride stress. J Food Agric Environ 4:240–243
Plomion C, Lalanne C, Claverol S, Meddour H et al (2006) Mapping the proteome of poplar and application to the discovery of drought-stress responsive proteins. Proteomics 6:6509–6527
Qian Y, Wilhelm SJ, Marcum KB (2001) Comparative responses of two Kentucky bluegrass cultivars to salinity stress. Crop Sci 41:1895–1900
Ramagopal S (1987) Salinity stress induced tissue-specific proteins in barley seedlings. Plant Physiol 84:324–331
Riccardi F, Gazeau P, Vienne D, Zivy M (1998) Protein changes in response to progressive water deficit in maize: quantitative variation and polypeptide identification. Plant Physiol 117:1253–1263
Rosen H (1957) A modified ninhydrin colorimetric analysis for amino acid. Arch Biochem Biophys 67:10–15
Roth U, Roepenack-Lahaye E, Clemens S (2006) Proteome changes in Arabidopsis thaliana roots upon exposure to Cd2+. J Exp Bot 57:4003–4013
Russell DA, Wong DML, Sachs MM (1970) The anaerobic response of soybean. Plant Physiol 92:401–407
Sairam RK, Tyagi A (2004) Physiology and molecular biology of salinity stress tolerance in plants. Curr Sci 86:407–421
Salekdeh GH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J (2002) Proteomic analysis of rice leaves during drought stress and recovery. Proteomics 2:1131–1145
Sanda S, Leustek T, Theisen MJ, Garavito RM, Benning C (2001) Recombinant Arabidopsis SQD1 converts UDP-glucose and sulfite to the sulfolipid head group precursor UDP-sulfoquinovose in vitro. J Biol Chem 276:3941–3946
Sappl PG, Onate-Sanchez L, Singh KB, Millar AH (2004) Proteomic analysis of glutathione S-transferases of Arabidopsis thaliana reveals differential salicylic acid-induced expression of the plant-specific phi and tau classes. Plant Mol Biol 54:05–219
Sarnighausen E, Wurtz V, Heintz D, Dorsselaer AV, Resk R (2004) Mapping of the Physcomitrella patens proteome. Phytochemistry 65:1589–1607
Seki M, Narusaka M, Ishida J, Nanjo T et al (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31:279–292
Sheffield J, Taylor N, Fauquet C, Chen S (2006) The cassava (Manihot esculenta Crantz) root proteome: protein identification and differential expression. Proteomics 6:1588–1598
Sioud M, Jespersen L (1996) Enhancement of hammerhead ri-bozyme catalysis by glyceraldehyde-3-phosphate dehydrogenase. J Mol Biol 257:775–789
Smith AP, DeRidder BP, Guo WJ, Seeley EH et al (2004) Proteomic analysis of Arabidopsis glutathione S-transferases from benoxacor- and copper-treated seedlings. J Biol Chem 279:26098–26104
Tonge R, Shaw J, Middleton B, Rowlinson R, Rayner S, Young J, Pognan F, Hawkins E, Currie I, Davison M (2001) Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics 1:377–396
Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14
Wang M, Peng Z, Li C, Li F et al (2008) Proteomic analysis on a high salt tolerance introgression strain of Triticum aestivum/Thinopyrum ponticum. Proteomics 8:1470–1489
Watson BS, Asirvatham VS, Wang L, Sumner LW (2003) Mapping the proteome of barrel medic (Medicago truncatula). Plant Physiol 131:1104–1123
Xu C, Huang B (2008) Proteomic response to heat stress in the roots of Agrostis grass species contrasting in heat tolerance. J Exp Bot 59:4183–4194
Xu C, Garrett WM, Sullivan JH, Caperna TJ, Natarajan S (2006) Separation and identification of soybean leaf proteins by two-dimensional gel electrophoresis and mass spectrometry. Phytochemistry 67:2431–2440
Xu C, Sullivan JH, Garrett WM, Caperna TJ, Natarajan S (2008a) Impact of solar ultraviolet-B on the proteome in soybean lines differing in flavonoid contents. Phytochemistry 69:38–48
Xu C, Xu Y, Huang B (2008b) Protein extraction for 2-dimensional gel electrophoresis of proteomic profiling in turfgrass. Crop Sci 48:1608–1614
Yan S, Tang Z, Su W, Sun W (2005) Proteomic analysis of salt stress-responsive proteins in rice roots. Proteomics 5:235–244
Yang Y, Kwon HB, Peng HP, Shih MC (1993) Stress responses and metabolic regulation of glyceraldehyde-3-phosphate dehydrogenase genes in Arabidopsis. Plant Physiol 101:209–216
Yang Q, Wang Y, Zhang J, Shi W (2007) Identification of aluminum-responsive proteins in rice roots by a proteomic approach: cysteine synthase as a key player in Al response. Proteomics 7:737–749
Zhang XP, Glaser E (2002) Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends Plant Sci 7:14–21
Zhou S, Sauve R, Thannhauser TW (2009) Proteome changes induced by aluminium stress in tomato roots. J Exp Bot 60:1849–1857
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6:66–71
Acknowledgments
The authors would like to thank Emily Merewitz, Yan Xu, and Longxing Hu for critical review of the manuscript. Our thanks are also due to the Rutgers Center of Turfgrass Science for funding support of this project.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Zou.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, C., Sibicky, T. & Huang, B. Protein profile analysis of salt-responsive proteins in leaves and roots in two cultivars of creeping bentgrass differing in salinity tolerance. Plant Cell Rep 29, 595–615 (2010). https://doi.org/10.1007/s00299-010-0847-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-010-0847-3