Abstract
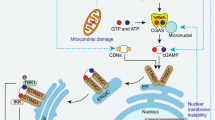
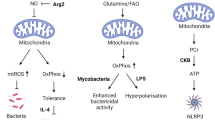
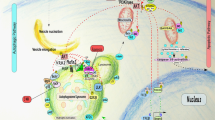
Autophagy is an essential, homeostatic process by which cells break down their own components. Perhaps the most primordial function of this lysosomal degradation pathway is adaptation to nutrient deprivation. However, in complex multicellular organisms, the core molecular machinery of autophagy — the 'autophagy proteins' — orchestrates diverse aspects of cellular and organismal responses to other dangerous stimuli such as infection. Recent developments reveal a crucial role for the autophagy pathway and proteins in immunity and inflammation. They balance the beneficial and detrimental effects of immunity and inflammation, and thereby may protect against infectious, autoimmune and inflammatory diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Deretic, V. & Levine, B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5, 527–549 (2009).
Virgin, H. W. & Levine, B. Autophagy genes in immunity. Nature Immunol. 10, 461–470 (2009).
Kroemer, G., Marino, G. & Levine, B. Autophagy and the integrated stress response. Mol. Cell 40, 280–293 (2010).
Saitoh, T. & Akira, S. Regulation of innate immune responses by autophagy-related proteins. J. Cell Biol. 189, 925–935 (2010).
Levine, B. & Kroemer, G. Autophagy in the pathogenesis of disease. Cell 132, 27–42 (2008).
Mizushima, N., Yoshimori, T. & Levine, B. Methods in mammalian autophagy research. Cell 140, 313–326 (2010). This paper provides a concise and critical review of current methods to monitor and modulate autophagy in mammalian cells.
Schmid, D., Pypaert, M. & Munz, C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity 26, 79–92 (2007).
Yu, L. et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465, 942–946 (2010).
Nakatogawa, H., Suzuki, K., Kamada, Y. & Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nature Rev. Mol. Cell Biol. 10, 458–467 (2009).
Hayashi-Nishino, M. et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nature Cell Biol. 11, 1433–1437 (2009).
Yla-Antilla, P., Vihinen, H., Jokitalo, E. & Eskelinen, E. L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5, 1180–1185 (2009).
Mizushima, N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22, 132–139 (2010).
Itakura, E. & Mizushima, N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6, 764–776 (2010).
Matsunaga, K. et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nature Cell Biol. 11, 385–396 (2009).
Axe, E. L. et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701 (2008).
Polson, H. E. et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 6, 506–522 (2010).
Ropolo, A. et al. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J. Biol. Chem. 282, 124–133 (2007).
Tian, Y. et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 141, 1042–1055 (2010).
Fujita, N. et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 19, 2092–2100 (2008).
Weidberg, H. et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29, 1792–1802 (2010).
Behrends, C., Sowa, M. E., Gygi, S. P. & Harper, J. W. Network organization of the human autophagy system. Nature 466, 68–76 (2010).
Radoshevich, L. et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 142, 590–600 (2010).
Saitoh, T. et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl Acad. Sci. USA 106, 20842–20846 (2009).
Webber, J. L. & Tooze, S. A. New insights into the function of Atg9. FEBS Lett. 584, 1319–1326 (2010).
Hailey, D. W. et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141, 656–667 (2010).
Ravikumar, B., Moreau, K., Jahreiss, L., Puri, C. & Rubinsztein, D. C. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nature Cell Biol. 12, 747–757 (2010).
English, L. et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nature Immunol. 10, 480–487 (2009).
Kraft, C., Peter, M. & Hofmann, K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nature Cell Biol. 12, 836–841 (2010).
Yamaguchi, H. et al. An initial step of GAS-containing autophagosome-like vacuoles formation requires Rab7. PLoS Pathogens 5, e1000670 (2009).
Nakagawa, I. et al. Autophagy defends cells against invading group A Streptococcus . Science 306, 1037–1040 (2004). Reference 30, together with reference 99, provides the first evidence that autophagy has a key role in bacterial infection. Atg5 is shown to be essential for controlling the replication of group A Streptococci that escape into the cytoplasm.
Sanjuan, M. A. et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257 (2007).
Lee, H. K. et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity 32, 227–239 (2010). This study shows that the autophagy machinery is necessary for dendritic cells to process and present extracellular microbial antigens for MHC class II presentation in vivo , which protects mice against lethal viral infection.
Berger, S. B. et al. SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages. Nature Immunol. 11, 920–927 (2010).
Huang, J. et al. Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl Acad. Sci. USA 106, 6226–6231 (2009).
Zhao, Z. et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4, 458–469 (2008).
Khaminets, A. et al. Coordinated loading of IRG resistance GTPases on to the Toxoplasma gondii parasitophorous vacuole. Cell. Microbiol. 12, 939–961 (2010).
Zhao, Y. O., Khaminets, A., Hunn, J. P. & Howard, J. C. Disruption of the Toxoplasma gondii parasitophorous vacuole by IFNγ-inducible immunity-related GTPases (IRG proteins) triggers necrotic cell death. PLoS Pathogens 5, 1–17 (2009).
Sumpter, R. Jr & Levine, B. Autophagy and innate immunity: triggering, targeting and tuning. Semin. Cell Dev. Biol. 21, 699–711 (2010).
Shahnazari, S. et al. A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe 8, 137–146 (2010).
Orvedahl, A. et al. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 7, 115–127 (2010).
Dupont, N. et al. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe 6, 137–149 (2009).
Ponpuak, M. et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 32, 329–341 (2010).
Guillemain, A. & Plus, N. Contrôle génique de la multiplication du virus de la sensibilité héréditaire au CO2 chez Drosophila melanogaster . Caryologia (suppl.) 1211–1213 (1954).
Mizushima, N. & Levine, B. Autophagy in mammalian development and differentiation. Nature Cell Biol. 12, 823–830 (2010).
Liu, Y. et al. Autophagy regulates programmed cell death during the plant innate immune response. Cell 121, 567–577 (2005).
Shelly, S., Lukinova, N., Bambina, S., Berman, A. & Cherry, S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 30, 588–598 (2009).
Yano, T. et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nature Immunol. 9, 908–916 (2008).
Jia, K. et al. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc. Natl Acad. Sci. USA 106, 14564–14569 (2009).
Singh, S. B., Davis, A. S., Taylor, G. A. & Deretic, V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 313, 1438–1441 (2006).
Intemann, C. D. et al. Autophagy gene variant IRGM –261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathogens 5, e1000577 (2009).
Kumar, D. et al. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis . Cell 140, 731–743 (2010).
Cooney, R. et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nature Med. 16, 90–97 (2010).
Travassos, L. H. et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nature Immunol. 11, 55–62 (2010).
Zhang, F. R. et al. Genomewide association study of leprosy. N. Engl. J. Med. 361, 2609–2618 (2009).
Dreux, M. & Chisari, F. V. Viruses and the autophagy machinery. Cell Cycle 9, 1295–1307 (2010).
Blanchet, F. P. et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity 32, 654–669 (2010).
Orvedahl, A. et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1, 23–35 (2007).
Leib, D. A., Alexander, D. E., Cox, D., Yin, J. & Ferguson, T. A. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T cell responses. J. Virol. 83, 12164–12171 (2009).
E. X. et al. Viral Bcl-2-mediated evasion of autophagy aids chronic infection of γherpesvirus 68. PLoS Pathogens 5, e1000609 (2009).
Lee, J. S. et al. FLIP-mediated autophagy regulation in cell death control. Nature Cell Biol. 11, 1355–1362 (2009).
Yoshikawa, Y. et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nature Cell Biol. 11, 1233–1240 (2009).
Moreau, K. et al. Autophagosomes can support Yersinia pseudotuberculosis replication in macrophages. Cell. Microbiol. 12, 1108–1123 (2010).
Kyei, G. B. et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 186, 255–268 (2009).
Dreux, M., Gastaminza, P., Wieland, S. F. & Chisari, F. V. The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl Acad. Sci. USA 106, 14046–14051 (2009). This study shows that many autophagy proteins are essential for the initial stages of hepatitis C viral RNA translation, illustrating that autophagy proteins may be subverted to enhance the replication of intracellular pathogens.
Sir, D. et al. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc. Natl Acad. Sci. USA 107, 4383–4388 (2010).
Reggiori, F. et al. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7, 500–508 (2010).
Yanai, H. et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature 462, 99–103 (2009).
Tang, D. et al. Endogenous HMGB1 regulates autophagy. J. Cell Biol. 190, 881–892 (2010).
Yuk, J. M. et al. Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe 6, 231–243 (2009).
Low, D. H. et al. A novel human tectonin protein with multivalent β-propeller folds interacts with ficolin and binds bacterial LPS. PLoS ONE 4, e6260 (2009).
Lipinski, M. M. et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev. Cell 18, 1041–1052 (2010).
Liu, F. et al. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood 116, 4806–4814 (2010).
Nedjic, J., Aichinger, M., Emmerich, J., Mizushima, N. & Klein, L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature 455, 396–400 (2008). This study provided the first evidence that the autophagy machinery functions in MHC class II antigen presentation in vivo , specifically in shaping the CD4+ T-cell repertoire during negative thymic selection, and thereby preventing autoimmunity and multi-organ inflammation.
Paludan, C. et al. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 307, 593–596 (2005). This study provided the first evidence that the autophagy machinery can deliver endogenously synthesized antigens for presentation on MHC class II molecules to CD4+ T cells.
Munz, C. Antigen processing via autophagy—not only for MHC class II presentation anymore? Curr. Opin. Immunol. 22, 89–93 (2010).
Lee, H. K., Lund, J. M., Ramanathan, B., Mizushima, N. & Iwasaki, A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 315, 1398–1401 (2007).
Tal, M. C. et al. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl Acad. Sci. USA 106, 2770–2775 (2009).
Jounai, N. et al. The Atg5–Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl Acad. Sci. USA 104, 14050–14055 (2007).
Moscat, J. & Diaz-Meco, M. T. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 137, 1001–1004 (2009).
Komatsu, M. et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature Cell Biol. 12, 213–223 (2010).
Cadwell, K. et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008).
Saitoh, T. et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 456, 264–268 (2008). This study demonstrates that autophagy proteins negatively control endotoxin-induced inflammasome activation. References 81 and 82 also show that autophagy gene deficiency increases susceptibility to experimentally induced inflammatory bowel disease.
Meissner, F., Molawi, K. & Zychlinsky, A. Mutant superoxide dismutase 1-induced IL-1β accelerates ALS pathogenesis. Proc. Natl Acad. Sci. USA 107, 13046–13050 (2010).
Qu, X. et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 128, 931–946 (2007). This paper shows that autophagy genes are necessary for apoptotic cells to generate engulfment signals required for successful apoptotic corpse clearance and the prevention of tissue inflammation.
Barrett, J. C. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nature Genet. 40, 955–962 (2008).
Brain, O., Allan, P. & Simmons, A. NOD2-mediated autophagy and Crohn disease. Autophagy 6, 412–414 (2010).
Lapaquette, P., Glasser, A. L., Huett, A., Xavier, R. J. & Darfeuille-Michaud, A. Crohn's disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell. Microbiol. 12, 99–113 (2010).
Kuballa, P., Huett, A., Rioux, J. D., Daly, M. J. & Xavier, R. J. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS ONE 3, e3391 (2008).
Fujita, N. et al. Differential involvement of Atg16L1 in Crohn disease and canonical autophagy: analysis of the organization of the Atg16L1 complex in fibroblasts. J. Biol. Chem. 284, 32602–32609 (2009).
Cadwell, K. et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell 141, 1135–1145 (2010). This study shows that both hypomorphic expression of an autophagy protein and a viral-infection trigger are necessary for experimentally induced inflammatory bowel disease, suggesting that the interaction between host defects in autophagy and environmental stressors such as infection may be crucial for the pathogenesis of certain inflammatory diseases.
Harley, J. B. et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nature Genet. 40, 204–210 (2008).
Gateva, V. et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nature Genet. 41, 1228–1233 (2009).
Han, J. W. et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nature Genet. 41, 1234–1237 (2009).
Nakamura, T. et al. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell 140, 338–348 (2010).
Yang, L., Li, P., Fu, S., Calay, E. S. & Hotamisligil, G. S. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 11, 467–478 (2010).
Rodriguez, A. et al. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 3, 211–222 (2006).
Hotamisligil, G. S. Inflammation and metabolic disorders. Nature 444, 860–867 (2006).
Luciani, A. et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nature Cell Biol. 12, 863–875 (2010).
Gutierrez, M. G. et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766 (2004). Reference 99, together with reference 30, provides the first evidence that autophagy has a key role in bacterial infection. Autophagy induction by IFN-γ, rapamycin or starvation results in the conversion of mycobacterial phagosomes into phagolysosomes, thereby enhancing mycobacterial killing.
Ogawa, M. et al. Escape of intracellular Shigella from autophagy. Science 307, 727–731 (2005).
Acknowledgements
The work in the authors' laboratories was supported by National Institutes of Health (NIH) grants RO1 CA109618 and U54 AI057156 (B.L.); by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and by the Takeda Science Foundation (N.M.); and by NIH grants RO1 AI054483, U54 AI057160, RO1 AI084887 and RO1 CA096511 and the Broad Medical Foundation (H.W.V.). We thank T. Stappenbeck for discussions, and A. Diehl and M. Harstein for scientific illustration. We apologize to those authors whose work could not be cited owing to space limitations.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reprints and permissions information is available at http://www.nature.com/reprints.
Rights and permissions
About this article
Cite this article
Levine, B., Mizushima, N. & Virgin, H. Autophagy in immunity and inflammation. Nature 469, 323–335 (2011). https://doi.org/10.1038/nature09782
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09782